PANCREATIC CANCER
The most common – but unfortunately also most serious – type of cancer in the pancreas is denoted pancreatic ductal adenoarcinoma. In this project, we aim at identifying and examining new molecular markers for this tumor type. The goal is to obtain a better understanding of how pancreatic ductal adenoarcinomas arise and grow. We have previously isolated the tumor cells by laser microdissection and analyzed them for mutations in the genes KRAS, BRAF and EGFR. Moreover, we studied the expression pattern of the postulated stem cell markers CD133 and CD44 in normal and cancerous pancreatic tissue. More recent projects have focused on how the gene ABO (determining blood type) may be implicated in pancreatic cancer and on the implications of those genetic alterations that are detected in pancreatic juice sampled directly from patients. Currently, we are aiming to develop a novel mouse model for pancreatic cancer based on mutations in the CEL gene, which encodes the digestive enzyme carboxyl ester lipase.
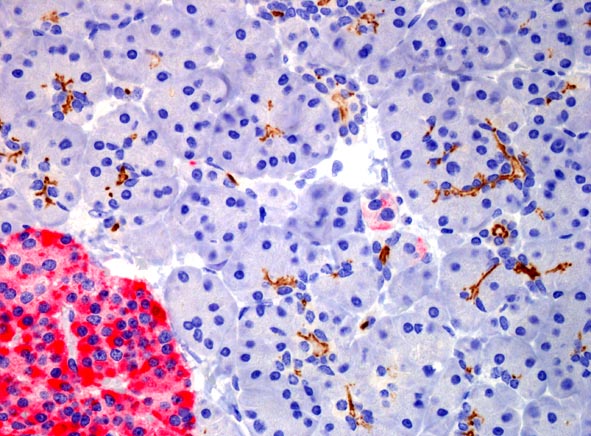
CD133 expression (brown) in normal pancreatic tissue.
The protein is expressed in acini centers and on the internal
surface of small ducts. The red area is an islet of Langerhans.
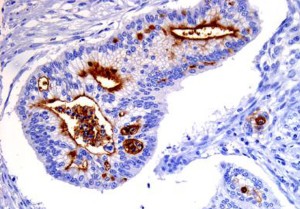
CD133 expression (brown) in pancreatic cancer. The
protein is strongly expressed on the internal surface
of duct-like structures formed by the cancer cells.
Current participants / major collaborators
Researcher Karianne Fjeld, Gade Laboratory for Pathology, UoBergen
PhD student Renate V. Seierstad, Gade Laboratory for Pathology, UoBergen
Master student Silje H. Johansen, Gade Laboratory for Pathology, UoBergen
Senior technician Solrun Steine, Gade Laboratory for Pathology, UoBergen
Dr. Marjolein Liedenbaum, Department of Radiology, Haukeland Univ. Hospital
Professor Ingfrid S. Haldorsen, Department of Clinical Medicine, UoBergen
Researcher Ivonne Regel, Dept. of Medicine, University Hospital LMU Munich, Germany
Professor Nils Halberg, Department of Biomedicine, UoBergen
Professor Oddmund Nordgård, Stavanger University Hospital/University of Stavanger
Professor Caroline S. Verbeke, Department of Pathology, University of Oslo
From 2020, we are part of the National Group of Expertise for Research on Pancreatic Cancer, funded by the Norwegian Cancer Society and led by Professor Caroline S. Verbeke, University of Oslo.
Publications
Brekke RS, Gravdal A, El Jellas K, Curry GE, Lin J, Wilhelm SJ, Steine SJ, Mas E, Johansson S, Lowe ME, Johansson BB, Xiao X, Fjeld K & Molven A (2024). Common single-base insertions in the VNTR of the carboxyl ester lipase (CEL) gene are benign and also likely to arise somatically in the exocrine pancreas. Human Molecular Genetics 33: 1001-1014.
Musiime M, Erusappan PM, Cukierman E, Chang J, Molven A, Hansen U, Zeltz C & Gullberg D (2024). Fibroblast integrin α11β1 is a collagen assembly receptor in mechanoregulated fibrillar adhesions. Matrix Biology 134: 144-161.
Tjensvoll K, Lapin M, Gilje B, Garresori H, Oltedal S, Forthun RB, Molven A, Rozenholc Y & Nordgård O (2022). Novel hybridization- and tag-based error-corrected method for sensitive ctDNA mutation detection using ion semiconductor sequencing. Scientific Reports 12: 5816.
Liu XZ, Rulina A, Choi MH, Pedersen L, Lepland J, Takle ST, Madeleine N, Peters SD, Wogsland CE, Grøndal SM, Lorens JB, Goodarzi H, Lønning PE, Knappskog S, Molven A & Halberg N (2022). C/EBPB-dependent adaptation to palmitic acid promotes stemness in hormone receptor negative breast cancer. Nature Communications 13: 69.
Choi MH, Tjora E, Forthun RB, Engjom T, Ræder H, Hovland R & Molven A (2021). KRAS mutation analysis by droplet digital PCR of duodenal juice from patients with MODY8 and other pancreatic diseases. Pancreatology 21: 1460-1465.
Shafiee S, Gelebart P, Popa M, Hellesøy M, Hovland R, Forthun RB, Lee J, Tohyama K, Molven A, Parekkadan B, Gjertsen BT, Kittang AO & McCormack E (2021). Preclinical characterisation and development of a novel myelodysplastic syndrome-derived cell line. British Journal of Haematology 193: 415-419.
Pedersen L, Panahandeh P, Siraji M, Knappskog S, Lønning PE, Zhu Q, Gordillo R, Scherer P, Molven A, Teigen K & Halberg N (2020). Golgi-localized PAQR4 mediates anti-apoptotic ceramidase activity in breast cancer. Cancer Research 80: 2163-2174.
Choi MH, Mejlænder-Andersen E, Manueldas S, El Jellas K, Steine SJ, Tjensvoll K, Sætran HA, Knappskog S, Hoem D, Nordgård O, Hovland R & Molven A (2019). Mutation analysis by deep sequencing of pancreatic juice from patients with pancreatic ductal adenocarcinoma. BMC Cancer 19: 11.
Zeltz C, Alam J, Liu H, Erusappan PM, Hoschuetzky H, Molven A, Parajuli H, Cukierman E, Costea DE, Lu N & Gullberg D (2019). α11β1 integrin is induced in a subset of cancer-associated fibroblasts in desmoplastic tumor stroma and mediates in vitro cell migration. Cancers (Basel) 11: 6.
El Jellas K, Johansson BB, Fjeld K, Antonopoulos A, Immervoll H, Choi MH, Hoem D, Lowe ME, Lombardo D, Njølstad PR, Dell A, Mas E, Haslam SM & Molven A (2018). The mucinous domain of pancreatic carboxyl-ester lipase (CEL) contains core 1/core 2 O-glycans that can be modified by ABO blood group determinants. Journal of Biological Chemistry 293:19476-19491.
El Jellas K, Hoem D, Hagen KG, Kalvenes MB, Aziz S, Steine SJ, Immervoll H, Johansson S, Molven A (2017). Associations between ABO blood groups and pancreatic ductal adenocarcinoma: influence on resection status and survival. Cancer Medicine 6: 1531-1540.
Dalva M, El Jellas K, Steine SJ, Ringdal M, Torsvik J, Immervoll H, Lerch MM, Johansson BB, Hoem D, Johansson S, Njølstad PR, Weiss U, Fjeld K & Molven A (2017). Copy number variants and VNTR length polymorphisms of the carboxyl-ester lipase (CEL) gene as risk factors in pancreatic cancer. Pancreatology 17: 83-88.
Calatayud D, Dehlendorff C, Boisen MK, Hasselby JP, Schultz NA, Werner J, Immervoll H, Molven A, Hansen CP & Johansen JS (2017). Tissue microRNA profiles as diagnostic and prognostic biomarkers in patients with resectable pancreatic ductal adenocarcinoma and periampullary cancers. Biomarker Research 5: 8.
Pettersen K, Andersen S, Degen S, Tadini V, Grosjean J, Hatakeyama S, Tesfahun AN, Moestue S, Kim J, Nonstad U, Romundstad PR, Skorpen F, Sørhaug S, Amundsen T, Grønberg BH, Strasser F, Stephens N, Hoem D, Molven A, Kaasa S, Fearon K, Jacobi C & Bjørkøy G (2017). Cancer cachexia associates with a systemic autophagy-inducing activity mimicked by cancer cell-derived IL-6 trans-signaling. Scientific Reports 7: 2046.
Dimcevski G, Kotopoulis S, Bjånes T, Hoem D, Schjøtt J, Gjertsen BT, Biermann M, Molven A, Sørbye H, McCormack E, Postema M & Gilja OH (2016). A human clinical trial using ultrasound and microbubbles to enhance treatment of inoperable pancreatic cancer. Journal of Controlled Release 243: 172-181.
Hoem D, Straume O, Immervoll H, Akslen LA & Molven A (2013). Vascular proliferation is associated with survival in pancreatic ductal adenocarcinoma. APMIS 121: 1037-1046.
Immervoll H, Hoem D, Steffensen OJ, Miletic H & Molven A (2011). Visualization of CD44 and CD133 in normal pancreas and pancreatic ductal adenocarcinomas: Non-overlapping membrane expression in cell populations positive for both markers. Journal of Histochemistry & Cytochemistry 59: 441-455.
Hoem D, Jensen D, Steine S, Thorsen TE, Viste A & Molven A (2008). Clinicopathological characteristics and non-adhesive organ culture of insulinomas. Scandinavian Journal of Surgery 97: 42-49.
Immervoll H, Hoem D, Sakariassen PØ, Steffensen OJ & Molven A (2008). Expression of the “stem cell marker” CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC Cancer 8: 48.
Wang J, Sakariassen PØ, Tsinkalovsky O, Immervoll H, Bøe SO, Svendsen A, Prestegarden L, Røsland G, Thorsen F, Stuhr L, Molven A, Bjerkvig R & Enger PØ (2008). CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. International Journal of Cancer 122: 761-768.
Søreide K, Immervoll H & Molven A (2006). Pancreatic intraepithelial neoplasia – precursors to pancreatic cancer. Tidsskrift for Den Norske Lægeforening 126: 905-908. [in Norwegian]
Immervoll H, Hoem D, Kugarajh K, Steine S & Molven A (2006). Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: Lack of mutations in the BRAF and EGFR genes. Virchows Archiv 448: 788-796.